Table of Contents
Introduction
The combined action of DNA regulatory elements and composition of locally bound proteins regulates gene expression. Integration of artificial reporter genes (as a sensor) into the genome is a powerful tool to explore the effects of regulatory elements and local chromatin context on gene expression. TRIP (short for thousands of reporters integrated in parallel) is a breakthrough technology that allows the parallel monitoring of thousands of integrated reporter genes throughout the genome.
Principle of TRIP
TRIP is based on a large library of reporter genes. These reporters are identical except for a short random DNA segment (16-20 bp), which we refer to as barcode. These reporters are randomly integrated throughout the genome in a pool of cells of interest. The reporters encounter different chromatin environments depending on their site of landing which changes their behavior accordingly. The barcode associated with each reporter serves as a unique tag for independent tracking of the reporter location and activity. 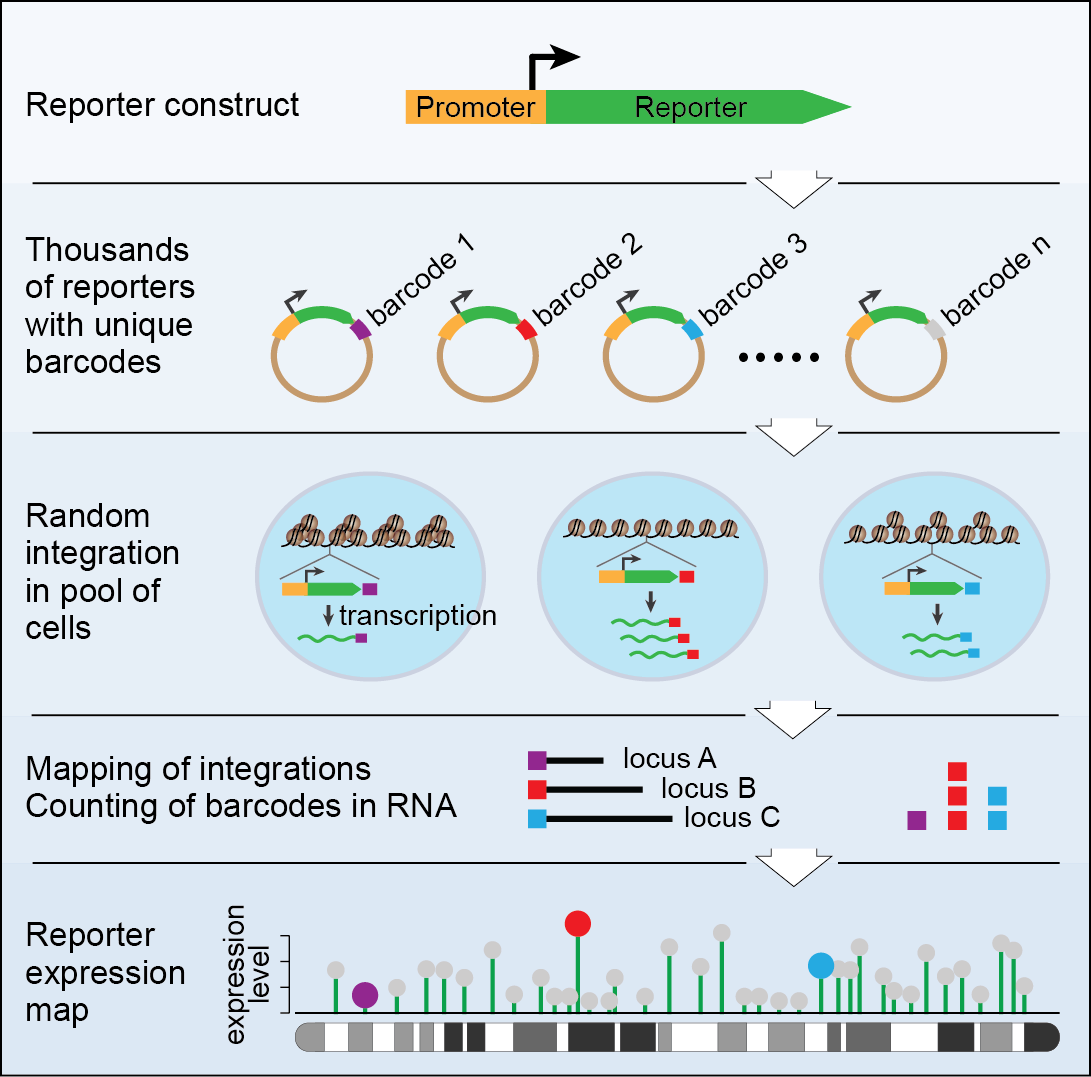 The parallel monitoring of thousands of integrated reporters provides a genome-wide atlas of reporter activity as a function of its position in the genome (Figure 1).
The parallel monitoring of thousands of integrated reporters provides a genome-wide atlas of reporter activity as a function of its position in the genome (Figure 1).
Insights gained through TRIP
A proof of principle experiment with a set of ~18,000 distinct reporter integrations in mouse embryonic stem cells has lead to novel insights into genome biology. Depending on their genomic position of integration, reporters show a surprisingly large (~1,000-fold) variation in their expression levels. This variation can be partially explained by whether a reporter is in a genomic region associated with nuclear lamina, by the degree of compaction of the chromatin, and by the proximity to promoters and enhancers of active endogenous genes. Another intriguing finding that came out of TRIP data is that neighboring active genes and enhancers generally influence gene expression on average over a distance of ~20 kb. Use of TRIP with an artificially inducible promoter has convincingly revealed that lamina associated domains repress transcription through an attenuation-based mechanism. Currently, TRIP is being used in our and many other labs to explore the impact of chromatin neighborhoods on a number of cellular processes related to gene expression.
Sequences of the TRIP plasmids
TRIP vector pPTK-Gal4-mPGK-Puro-IRES-eGFP-sNRP-pA
TRIP vector pPTK-Gal4-tet-Off-Puro-IRES-eGFP-sNRP-pA
TRIP vector pPTK-Gal4-tet-Off-Puro-IRES-eGFP-sNRP-pA-trim1
TRIP vector pPTK-Gal4-mPGK-Puro-IRES-eGFP-sNRP-pA-BC-Library
TRIP vector pPTK-Gal4-tet-Off-Puro-IRES-eGFP-sNRP-pA-BC-Library
The TRIP Analysis Software Kit (TASK)
For processing of raw sequencing data from TRIP experiments, we have developed a set of scripts. The scripts are tested on both Mac OS X and Ubuntu 12.04LTS.
trip
trip is a Perl and R based program to generate a table of barcode sequences, their counts and mapping information given fastq files for normalization, expression and mapping reads. The script depends on the programming language Perl and the Perl module Text::LevenshteinXS, the alignment tool Bowtie2, the programming language R and the R packages ggplot2 and gridExtra. For details see the user manual provided with the script.
Useful Linkes
TRIP Publications
Akhtar W*, de Jong J*, Pindyurin AV*, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M,, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel (2013). Cell 154(4):914-27 Journal site PubMed.
Akhtar W*, Pindyurin AV*, de Jong J, Pagie L, ten Hoeve J, Berns A, Wessels LF, van Steensel B, van Lohuizen M, Using TRIP for genome-wide position effect analysis in cultured cells (2014). Nature Protocols 9(6):1255–1281 Journal site PubMed
de Jong J, Wessels LF, van Lohuizen M, de Ridder J, Akhtar W, (2015). Applications of DNA integrating elements: Facing the bias bully. Mobile Genetic Elements 6:1-6 Journal site * These authors contributed equally to this work
Contacts
If you have any questions regarding TRIP and TRIP data analysis, please feel free to contact us.

